Mapped IDs
Include/exclude samples
Expression levels
Loading...
Missing values cutoff
Set maximum number allowed missing values for each treatment
To proceed without removing any proteins with missing values, set the cutoff to a value higher than the number of samples / treatments
To proceed without removing any proteins with missing values, set the cutoff to a value higher than the number of samples / treatments
Validate IDs
Please note: Depending on the number of IDs, this process may take a long time to run.
Convert IDs
Assess goodness-of-fit
Find closest human homologue using BLAST.
Generate report
Reproducibility token
PCA settings
Loading...
Heatmap settings
Sample-sample correlation heatmap
Loading...
Set contrasts
Which treatments should be compared?
Filter table by:
Loading...
Pathway settings
Selected pathway info
Loading...
Loading...
 ProteoMill
ProteoMill
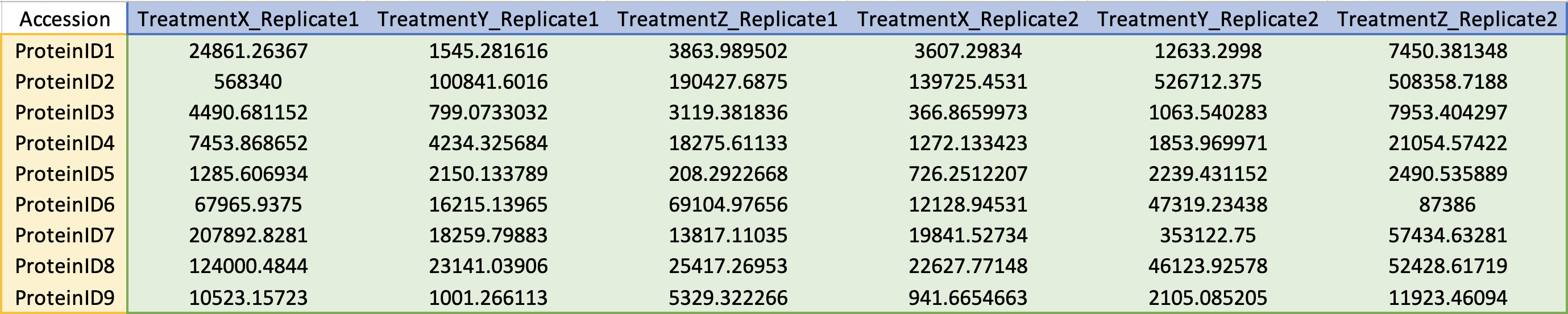
 Protein IDs.
In the first column, row two and onwards contains protein IDs (see accepted ID types below).
Protein IDs.
In the first column, row two and onwards contains protein IDs (see accepted ID types below).
 Sample names.
In the first row, column two and onward contains the sample names, with treatments and replicates separated by an underscore character. Sample column order does not matter.
Sample names.
In the first row, column two and onward contains the sample names, with treatments and replicates separated by an underscore character. Sample column order does not matter.
 Quantitative values.
Only numeric values greater than or equal to 0, or NA. Period (.) should be used for decimal character.
Quantitative values.
Only numeric values greater than or equal to 0, or NA. Period (.) should be used for decimal character.